Research Interests
- Fields: Biomedical Signal Processing; Computational Physiology and Neuroscience, Statistical Physics
- Research Activity: Development of advanced biomedical signal processing methods for the analysis of complex physiological systems, aimed at mechanism understanding and disease assessment
- Methodological approach: Measurement of physiological time series from biomedical signals; development of methods for multivariate time series analysis in the time domain (prediction methods), frequency domain (spectral analysis) and information domain (entropy-based measures) for the quantitative description of the complexity of individual systems, the coupling between systems and their causal interaction.
- Applicative contexts: neurophysiology; brain connectivity; cognitive neuroscience; cardiovascular neuroscience; cardiac, cardiorespiratory and cerebrovascular regulation; heart rate variability; cardiac atrial fibrillation; brain-heart interactions; network physiology.
- Aims: characterization of brain, cardiac and multi-organ physiological mechanisms in physiological states (e.g.: aging, sleep, cognition, resting states, physiological stressors) and diseased conditions (e.g.: sleep disorders, syncope, epilepsy, cardiac fibrillation).
|
Research Lines
Multi-System Analysis of the Human Physiological Network
Based on the view of the human body as an integrated network composed
by several organ systems, each with its own internal dynamics but also
functionally connected to the other organs, we apply novel methods for
multivariate time series analysis to the nonlinear, multi-scale and
time-variant output signals of brain, heart and peripheral
physiological systems. This integrated unconventional approach aims at
providing new insight on the functional structure of the human
physiological networks and on its evolution across different
physiological states and pathological conditions. A prime example of
application of this multi-system approach is the study of sleep state
transitions and sleep disorders.
- A Porta, L Faes, 'Wiener-Granger Causality in Network Physiology with Applications to Cardiovascular Control and Neuroscience', Proceedings of the IEEE 2016; 104(2): 282-309.
- L Faes, D Marinazzo, S Stramaglia, F Jurysta, A Porta, G Nollo, 'Predictability decomposition detects the impairment of brain-heart dynamical networks during sleep disorders and their recovery with treatment', Phil. Trans. R. Soc. A, special issue on "Uncovering brain-heart information through advanced signal and image processing", 2016; 374:20150177.
- L Faes, D Marinazzo, F Jurysta, G Nollo, 'Linear and nonlinear analysis of brain-heart and brain-brain interactions during sleep', Phys. Meas. 2015; 36:683-698.
------------------
Information Dynamics of Brain Networks
In order to improve the understanding
of how the brain function emerges from the coordinated behavior of
spatially separated cerebral regions, we study the temporal dynamics of
brain networks reconstructed from EEG or fMRI measurements. We define
entropy-based measures quantifying the amounts of information generated
at each network node, actively stored in the node, transferred to it
from the other connected nodes, and modified during the transfer
according to synergetic and/or redundant interactions.
Information-theoretic measures are complemented with frequency-domain
measures of brain connectivity to provide a complete analysis framework
that is exploited to assess resting-state and brain networks and their
modifications induced by stimulation or cognitive elicitation.
- L Faes, D Marinazzo, G Nollo, A Porta 'An information-theoretic framework to map the spatio-temporal dynamics of the scalp electroencephalogram', IEEE Trans. Biomed. Eng., special issue on “Brain Connectivity”, in press, 2016; DOI: 10.1109/TBME.2016.2569823.
- S Stramaglia, L Angelini, G Wu, JM Cortes, L Faes, D Marinazzo, 'Synergetic and redundant information flow detected by unnormalized Granger causality: application to resting state fMRI', IEEE Trans. Biomed. Eng., special issue on "Brain Connectivity", in press, 2016; DOI: 10.1109/TBME.2016.2559578.
- A Montalto, L Faes, D. Marinazzo, 'MuTE: a MATLAB toolbox to compare established and novel estimators of the multivariate transfer entropy', PLOS ONE 2014; 9(10):e109462 (13 pages).
------------------
Advanced Tools for Assessing the Complexity of Cardiovascular Regulation
The short-term analysis of heart
rate, arterial pressure and respiration beat-to-beat variability
provides a non-invasive quantitative way to assess how the autonomic
nervous system affects the cardiovascular control. We develop novel
time series analysis tools, mainly rooted in information theory, to
quantify the complexity of cardiac dynamics and how this complexity is
explained by the vascular and respiratory dynamics. These tools are
then exploited to investigate the mechanisms underlying cardiovascular
and cardiorespiratory regulation under different physiological
stressors and pathological states.
- M Javorka, B Czippelova, Z Turianikova, Z Lazarova, I Tonhajzerova, L Faes, 'Causal analysis of short-term cardiovascular variability: state-dependent contribution of feedback and feedforward mechanisms', Med. Biol. Eng. Comput., in press, 2016; DOI: 10.1007/s11517-016-1492-y.
- D Widjaja, A Montalto, E Vlemincx, D Marinazzo, S Van Huffel, L Faes, 'Cardiorespiratory information dynamics during mental arithmetic and sustained attention', PLOS ONE 2015; 10(6): e0129112 (14 pages).
- L Faes, D Marinazzo, A Montalto, G Nollo, 'Lag-specific transfer entropy as a tool to assess cardiovascular and cardiorespiratory information transfer', IEEE Trans Biomed Eng 2014; 61(10):2556-2568. DOI: 10.1109/TBME.2014.2323131
------------------
Filling the Gap between Advanced Biosignal Methods, Clinical Practice and Personalized Medicine
The exploitation of tools for
biomedical signal processing and time series analysis in the clinical
practice is currently limited, among other factors, by the lack of a
standardized assessment of descriptive measures, the computational
burden of their calculation, and the poor consideration of
inter-individual differences. Focusing on the evaluation of measures
which have shown their diagnostic power (e.g., complexity indexes of
heart rate variability), we face these issues by comparing a wide range
of analysis techniques, implementing data-efficient algorithms for
their computation, and designing statistical procedures to establish
the confidence limits of their estimates. Our goals are to achieve a
computational breakthrough in the assessment of complex physiological
dynamics, as well as to characterize individual differences in
cardiovascular control in a number of experimental conditions. This
will allow to provide a better knowledge on the specific physiology of
each individual, leading to a routinely feasible personalized
assessment of the underlying physiology, and ultimately to an improved
diagnosis of altered conditions.
- L Faes, DMS Simpson, A Beda, 'Estimation of Confidence Limits for Descriptive Indexes Derived from Autoregressive analysis of Time Series: Methods and Application to Heart Rate Variability', submitted, 2016.
- A Porta, B De Maria, V Bari, A Marchi, L Faes, 'Are Nonlinear Model-Free Approaches for the Assessment of the Entropy-Based Complexity of the Cardiac Control Superior to the Linear Model-Based one?', submitted, 2017.
- A Porta, G
Nollo, L Faes, 'Editorial: Bridging the gap between the development of
advanced biomedical signal processing tools and clinical practice',
Phys. Meas. 2015; 36:627-631.
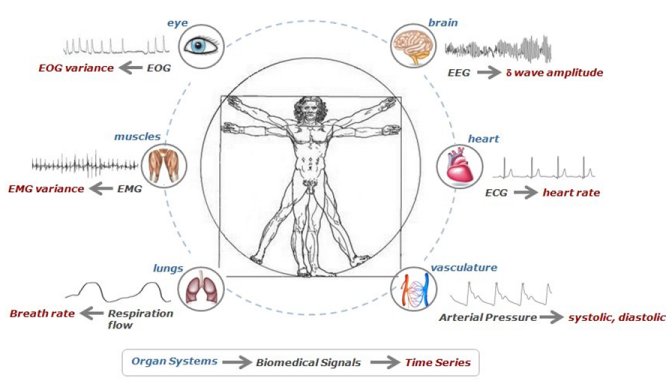
Network Physiology: how different organ systems dynamically interact
Last updated Jun 23, 2016